[NIPPONGENE] Thermostable Strand-Displacement DNA Polymerase
This product is an enzyme with 5' → 3' DNA polymerase activity and strand displacement activity, synthesizing a new DNA strand by releasing itself from the hydrogen bonds of double-stranded DNA that is used as a template. Thermostable strand-displacement DNA polymerases, which require no dissociation of double-stranded DNA due to their characteristics, can synthesize DNA at a constant temperature without being inhibited by the secondary structure of the DNA.
Features
- 5' → 3' DNA polymerase activity and strand displacement activity
- Able to synthesize DNA at a constant temperature
- Lineup of three enzymes that differ in reaction temperature
- Most suitable for synthesizing DNA strands with high GC content
Use
- Application based on strand displacement activity
(e.g., isothermal amplification)
- Application based on strand displacement activity
Product lineup
| Product name | Optimal temperature | Deactivation temperature*1 | Component | |
|---|---|---|---|---|
| BST DNA Polymerase | 60~65℃ | 80℃, 5 min | 1) BST DNA Polymerase (8units/μl) 2) 10× BST Reaction Buffer (80mmol/l Mg2+) |
1,600units 500μl |
| Csa DNA Polymerase | 60~70℃ | 85℃, 5 min | 1) Csa DNA Polymerase (8units/μl) 2) 10× Csa DNA Polymerase (80mmol/l Mg2+) |
1,600units 500μl |
| 96-7 DNA Polymerase | 50~55℃ | 70℃, 5 min | 1) 96-7 DNA Polymerase (8units/μl) 2) 10× 96-7 DNA Polymerase (95mmol/l Mg2+) |
1,600units 500μl |
*1 Inactivation temperature when enzyme stock solution is directly heat denatured.
≪Bst DNA Polymerase≫ Application for LAMP
LAMP reaction conditions
-
Bst DNA Polymerase
Reaction: 65°C, 60 min<Composition of reaction mixture>
Candidatus Liberibacter asiaticus DNA※2 1×106copies FIP 40pmol BIP 40pmol F3 Primer 5pmol B3 Primer 5pmol Loop Primer F 20pmol Loop Primer B 20pmol dNTPs Mixture 1.4mM each 10× Bst Reaction Buffer 2.5 μl Bst DNA Polymerase 8units Total 25μl *2 The LAMP primer set for detection of Candidatus Liberibacter asiaticus was developed by the Kyushu Okinawa Agricultural Research Center of the National Agriculture and Food Research Organization under a project titled "Development of technology to prevent the spread of pesticide-resistant citrus greening disease," one of their projects promoting agricultural forestry and fisheries research using advanced technology.
-
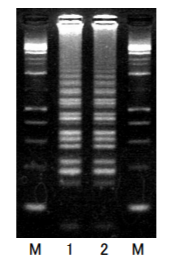
<Lane>
M:Gene-Ladder Wide 1(Product Number 313-06961)
1 : Company A Bst DNA Polymerase
2 : NIPPONGENE Bst DNA Polymerase<Note>
3.0% agarose 21/TAE gel electrophoresis
Ethidium bromide staining
Primer sequence
FIP: 5'-GCATGCCGAGGATCAATGCCTTGCTTAAAGAGCGTGCTACG-3'
BIP: 5'-TATGCCTAATGGCACGGGGGTAAGCTTCATCCGCCTTCGA-3‘
F3 Primer: 5'-TGGGTTAAGTGATGCTGTGG-3'
B3 Primer: 5'-CAACAATATCAGCCCCTGCT-3'
Loop Primer F: 5'-TCTCAACTGTTTCATCAAACCTAGC-3'
Loop Primer B: 5'- CGTGGCGGTTTTTGCTACA-3'
≪Csa DNA Polymerase / 96-7 DNA Polymerase≫ Application for LAMP
LAMP reaction conditions
<Composition of reaction mixture>
-
Csa DNA Polymerase
Candidatus Liberibacter asiaticus DNA 1×106copies FIP 40pmol BIP 40pmol F3 Primer 5pmol B3 Primer 5pmol Loop Primer F 20pmol Loop Primer B 20pmol dNTPs Mixture 1.4mM each 10× Csa Reaction Buffer 1× Csa DNA Polymerase 8units total 25μl Reaction: 65°C, 60 min
-
96-7 DNA Polymerase
Candidatus Liberibacter asiaticus DNA 1×106copies FIP 40pmol BIP 40pmol F3 Primer 5pmol B3 Primer 5pmol Loop Primer F 20pmol Loop Primer B 20pmol dNTPs Mixture 2.0mM each 10× 96-7 Reaction Buffer 1× 96-7 DNA Polymerase 8units total 25μl Reaction: 65°C, 60 min
Primer Sequence
-
FIP: 5'-GCATGCCGAGGATCAATGCCTTGCTTAAAGAGCGTGCTACG-3'
BIP: 5'-TATGCCTAATGGCACGGGGGTAAGCTTCATCCGCCTTCGA-3‘
F3 Primer: 5'-TGGGTTAAGTGATGCTGTGG-3'
B3 Primer: 5'-CAACAATATCAGCCCCTGCT-3'
Loop Primer F: 5'-TCTCAACTGTTTCATCAAACCTAGC-3'
Loop Primer B: 5'- CGTGGCGGTTTTTGCTACA-3' -
<Note>
3.0% agarose 21/TAE gel electrophoresis
Ethidium bromide staining
-
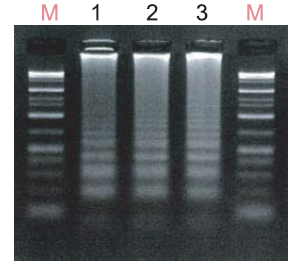
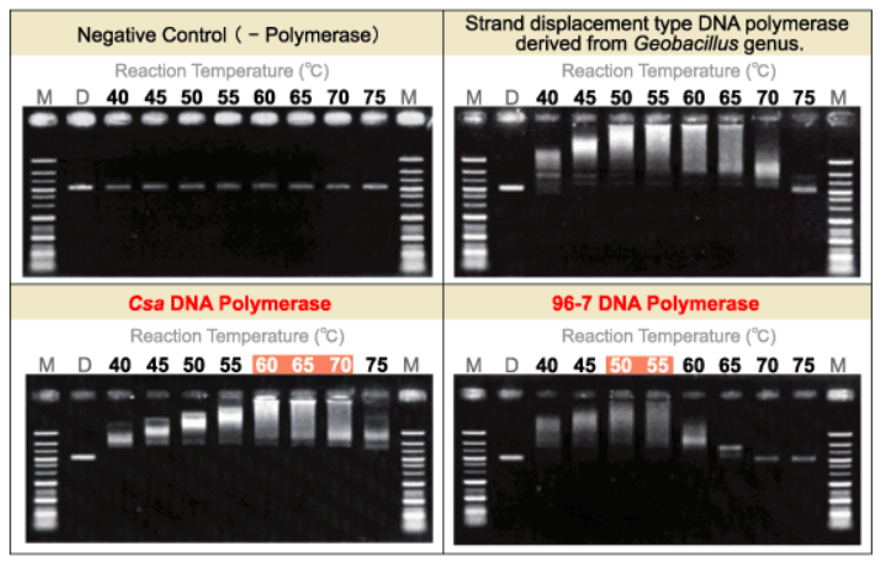
-
<Lane>
M: Gene-Ladder Wide 1(Product Number 313-06961)
1: Strand-displacement DNA polymerase derived from Geobacillus
2: NIPPONGENE Csa DNA Polymerase
3: NIPPONGENE 96-7 DNA Polymerase
≪Csa DNA Polymeras / 96-7 DNA Polymerase≫ Evaluation of thermostability and optimum temperature by RCA method
RCA reaction conditions
<Composition of reaction mixture>
-
M13mp18 single strand DNA 20ng Universal Primer 50nM dNTPs Mixture 0.25mM< each Tris-HCl (pH 8.8 at 25℃) 20mM KCl 10mM (NH4)2SO4 10mM MgSO4 2mM Tween 20 0.1% DNA Polymerase 8units total 20μl -
<Lane>
M: Gene-Ladder Wide 1(Product Number 313-06961)
D: Template DNA(M13mp18) single strand DNA
Primer Sequence
Universal Primer
5'-GTTTTCCCAGTCACGACGTTGTA-3'
<Note>
3.0% agarose 21/TAE gel electrophoresis
Ethidium bromide staining
RCA(Rolling Circle Amplification) reaction diagram

Product List
- Open All
- Close All
For research use or further manufacturing use only. Not for use in diagnostic procedures.
Product content may differ from the actual image due to minor specification changes etc.
If the revision of product standards and packaging standards has been made, there is a case where the actual product specifications and images are different.